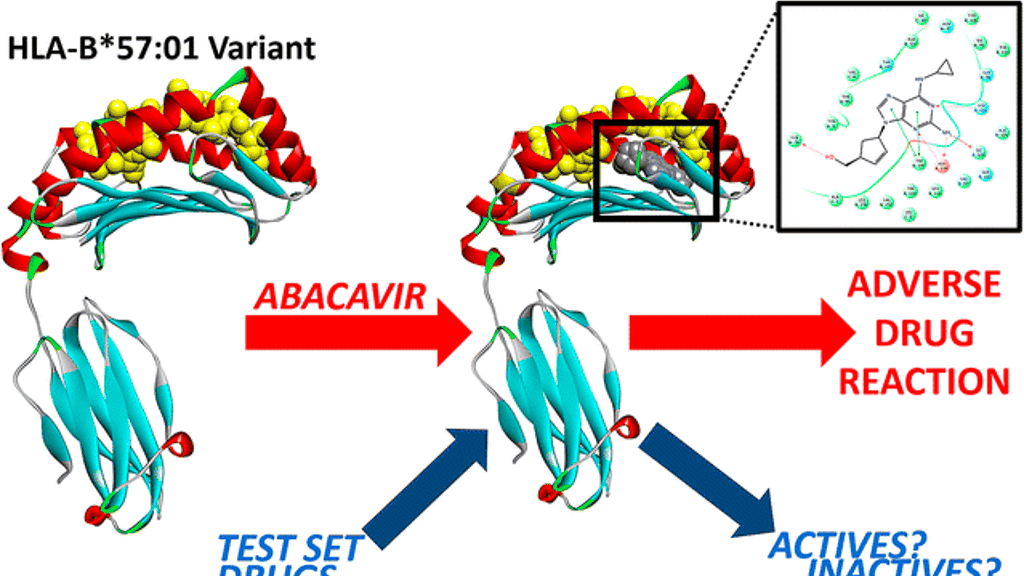
New computer models from North Carolina State University (NYSU) show how a variant of a common protein involved in human immune response binds to the antiviral drug abacavir, causing a severe life-threatening reaction known as the abacavir hypersensitivity syndrome (AHS). The work has implications for predicting severe adverse reactions caused by existing drugs and future drug candidates in subpopulations of patients.
Abacavir is a common anti-HIV drug. However, it is associated with severe allergic reactions in a fraction (5-8 percent) of patients who take it. Previous research determined that this response occurs in patients with a particular variant of a human leukocyte antigen (HLA) known as HLA-B*57:01. Denis Fourches, assistant professor of chemistry at NC State, and his graduate student, George Van Den Driessche, wanted to know what happens at the molecular level when abacavir and other drugs interact with HLA-B*57:01.
Their research has been published in the Journal of Cheminformatics, and was funded by the NC State Chancellor’s Faculty Excellence Program. Van Den Driessche is first author of the paper, Fourches is the corresponding author.
There are 15,000 variants of HLA, and everyone carries some of these variants. HLA-B*57:01 is one of the first variants studied in the context of a drug-induced immune reactions. It binds with abacavir, but little was understood about exactly what was happening structurally at the molecular level, especially in terms of the relationship with the co-binding peptide.
“The models allowed us to identify key atomic interactions that cause abacavir and other drugs to bind to the HLA variant protein and ultimately trigger the immune response,” Fourches explains. “When you can forecast and understand the elements of the drug that enable the binding to occur, you may be able to create new active compounds that do not have that problem.”
The researchers’ ultimate goal is to use molecular modeling to discover and understand how different drugs can interact with the immune system via direct HLA binding interaction, so that better predictions of the side-effects of new drugs and designing drugs with fewer side-effects becomes possible. This, the researchers believe, is critical for the personalized medicine of tomorrow.
Abacavir is a common anti-HIV drug. However, it is associated with severe allergic reactions in a fraction (5-8 percent) of patients who take it. Previous research determined that this response occurs in patients with a particular variant of a human leukocyte antigen (HLA) known as HLA-B*57:01. Denis Fourches, assistant professor of chemistry at NC State, and his graduate student, George Van Den Driessche, wanted to know what happens at the molecular level when abacavir and other drugs interact with HLA-B*57:01.
Their research has been published in the Journal of Cheminformatics, and was funded by the NC State Chancellor’s Faculty Excellence Program. Van Den Driessche is first author of the paper, Fourches is the corresponding author.
Triggering immune response
HLA proteins reside on the surface of cells. They help the immune system distinguish between its own proteins and those made by infectious agents. HLA proteins and their co-binding peptides serve as signaling proteins that communicate with T-cells, ensuring all is well. If something foreign – a pathogen or a drug like abacavir – binds to the HLA protein, displacing the co-binding peptide and deforming the overall shape of the molecular complex, this change is recognized by immune cells, triggering the immune response.There are 15,000 variants of HLA, and everyone carries some of these variants. HLA-B*57:01 is one of the first variants studied in the context of a drug-induced immune reactions. It binds with abacavir, but little was understood about exactly what was happening structurally at the molecular level, especially in terms of the relationship with the co-binding peptide.
Series of computer models
To better understand this complex system, Fourches and Van Den Driessche created a series of computer models using three-dimensional molecular docking that allowed them to look at the ways in which abacavir docks in the binding site of HLA-B*57:01 – where the co-binding peptide normally docks. The models incorporated 3D structures of abacavir, HLA-B*57:01, and several potential co-binding peptides. The researchers ran molecular modeling simulations to virtually dock abacavir in the HLA-B*57:01 active site in the absence and presence of a co-binding peptide. Finally, they virtually screened an additional set of 13 drugs, some of which are known to cause severe adverse responses that are also suspected of being linked to HLA variants.“The models allowed us to identify key atomic interactions that cause abacavir and other drugs to bind to the HLA variant protein and ultimately trigger the immune response,” Fourches explains. “When you can forecast and understand the elements of the drug that enable the binding to occur, you may be able to create new active compounds that do not have that problem.”
The researchers’ ultimate goal is to use molecular modeling to discover and understand how different drugs can interact with the immune system via direct HLA binding interaction, so that better predictions of the side-effects of new drugs and designing drugs with fewer side-effects becomes possible. This, the researchers believe, is critical for the personalized medicine of tomorrow.